
Alazygatr
alazygatr_vignette.RmdInstall all of the required packages:
Are you a lazy algatr? The do_everything_for_me()
function runs all six landscape genomic methods in the algatr package
(wingen, TESS, GDM, MMRR, RDA, and LFMM) and provides limited options
for customizability. This function primarily exists for fun and to
demonstrate that algatr really can be run using on a vcf and sampling
coordinates; we do not encourage researchers to
actually perform analyses on their data using this function!
The main arguments within this function are simple: vcf
specifies the vcf, coords specifies the sampling
coordinates, and envlayers specifies the environmental
layers (not required). As usual, make sure your samples are in the same
order between your data file and coordinates file and that your CRS is
consistent!
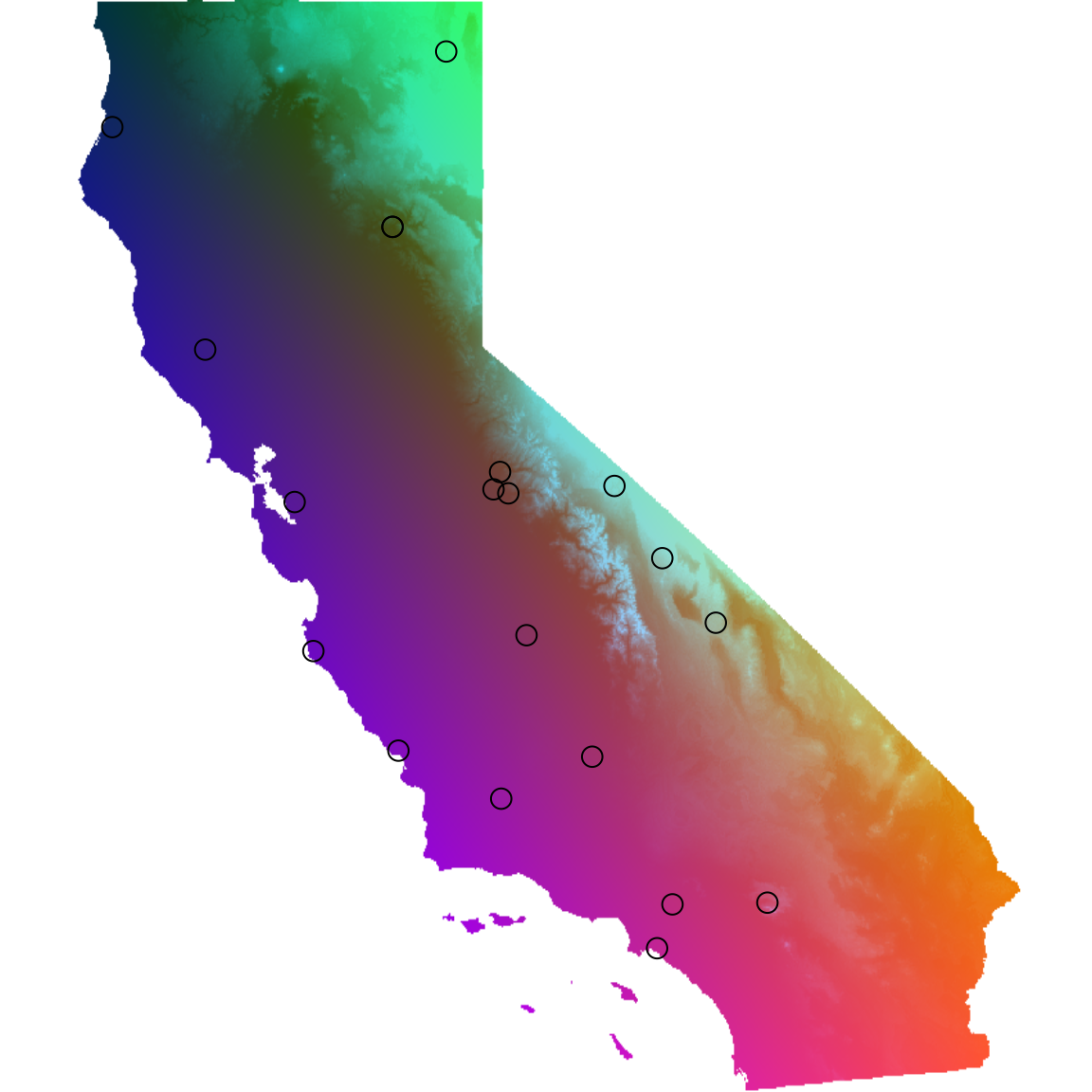
First, let’s load our test data. For the sake of things running quickly, let’s run this function on only 20 individuals from the test dataset.
##
## ---------------- example dataset ----------------
##
## Objects loaded:
## *liz_vcf* vcfR object (1000 loci x 53 samples)
## *liz_gendist* genetic distance matrix (Plink Distance)
## *liz_coords* dataframe with x and y coordinates
## *CA_env* RasterStack with example environmental layers
##
## -------------------------------------------------
## ##
gen <- liz_vcf[, 1:21]## Loading required namespace: vcfR
coords <- liz_coords[1:20, ]
envlayers <- CA_envNow, let’s run the function:
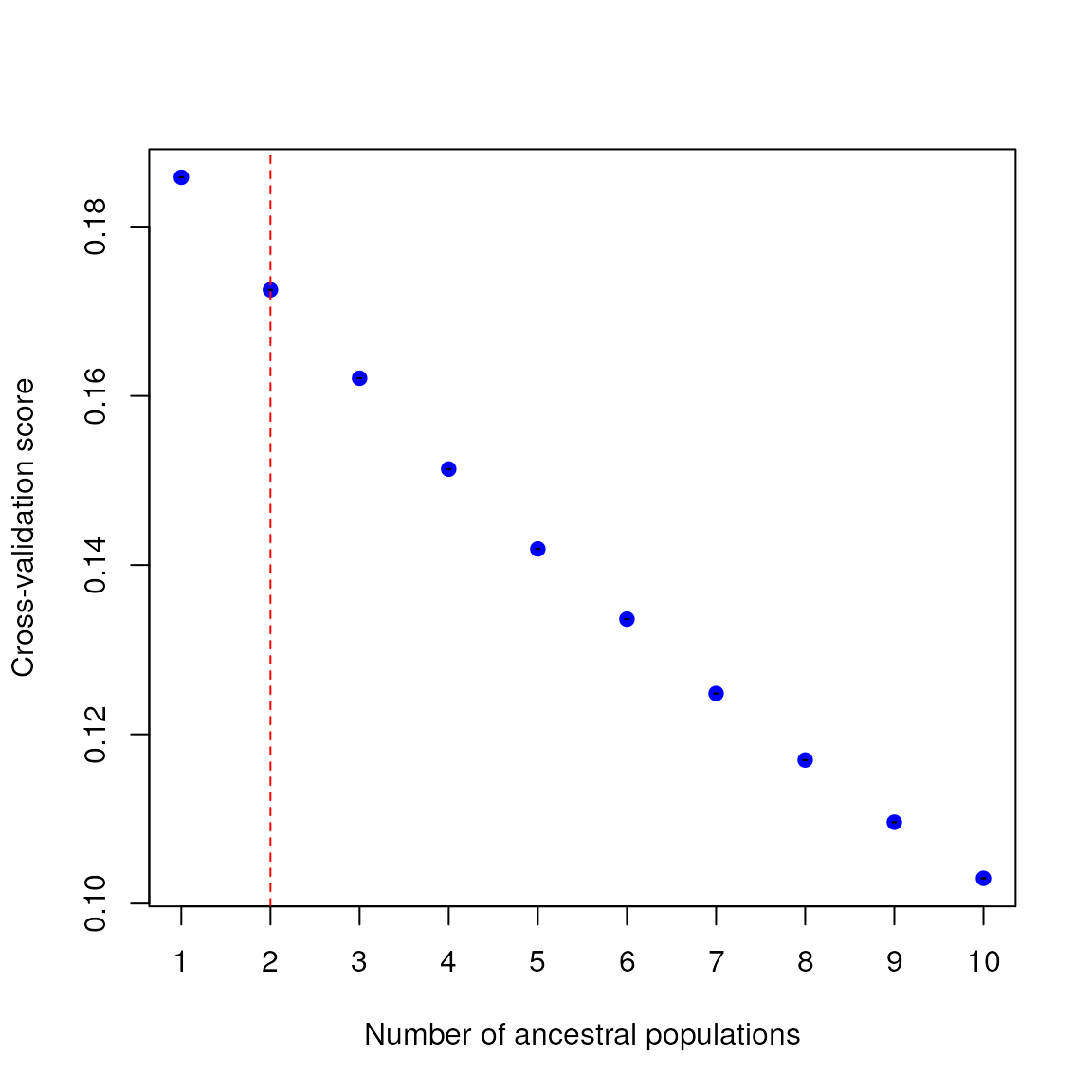
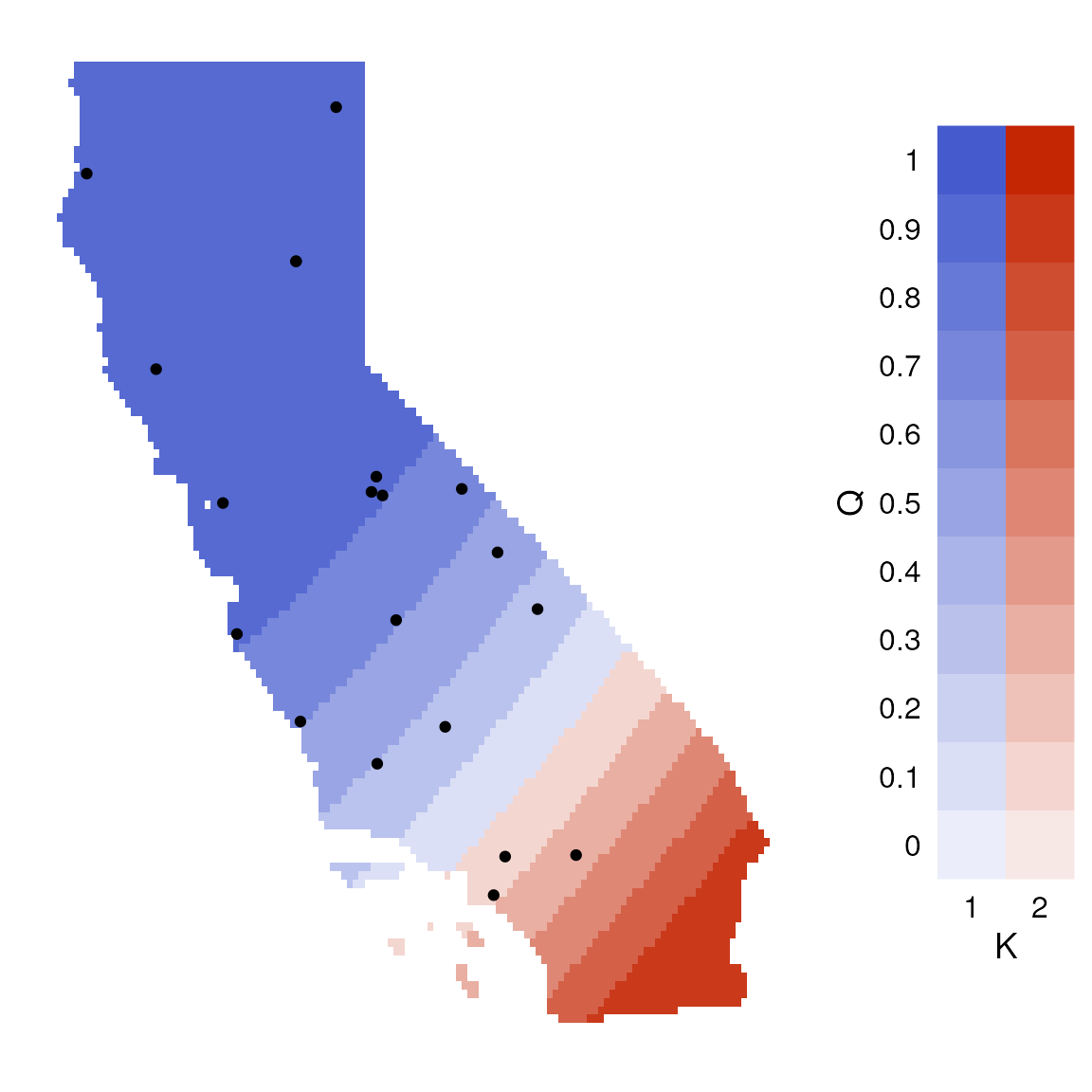
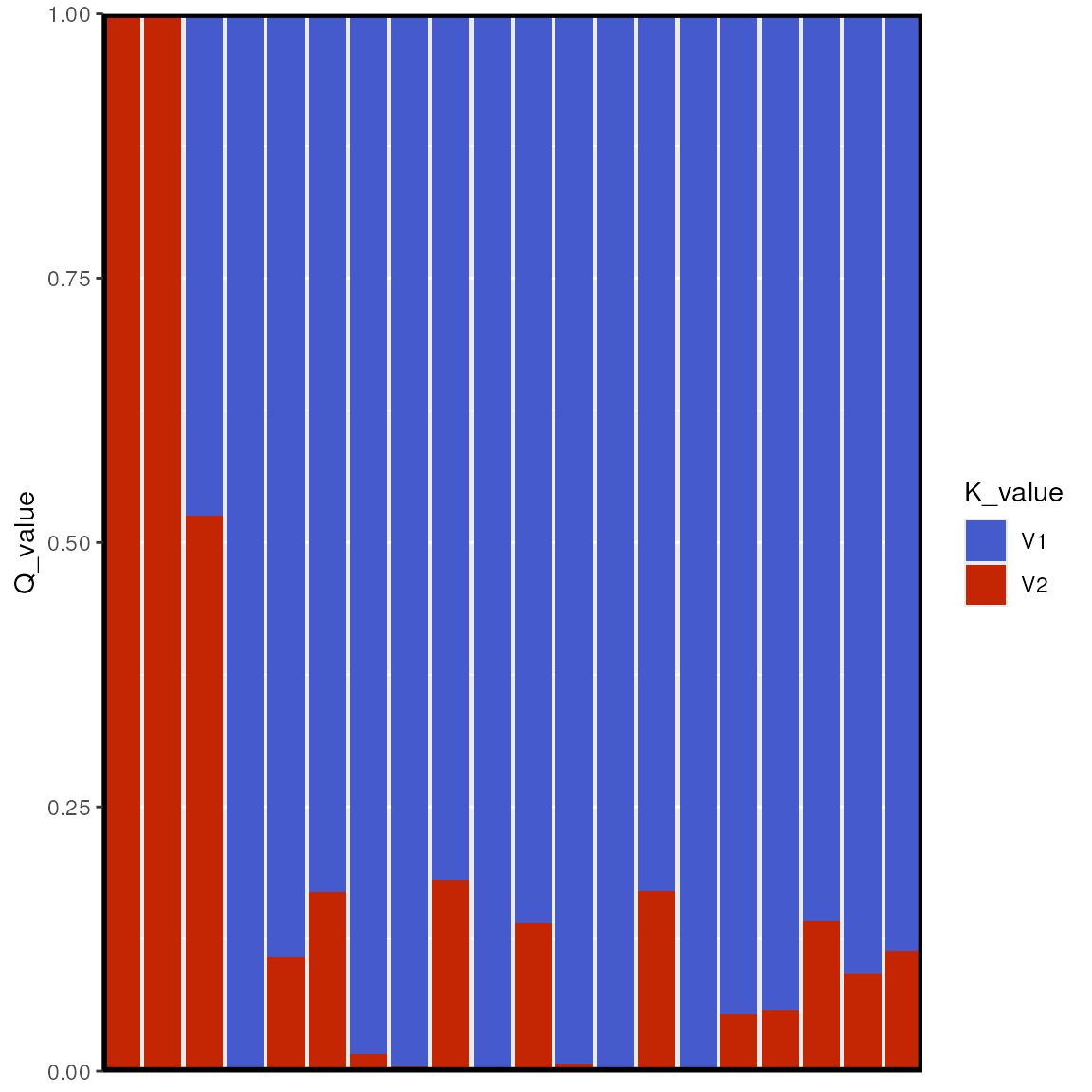
lazy_results <- do_everything_for_me(gen, coords, envlayers, quiet = FALSE)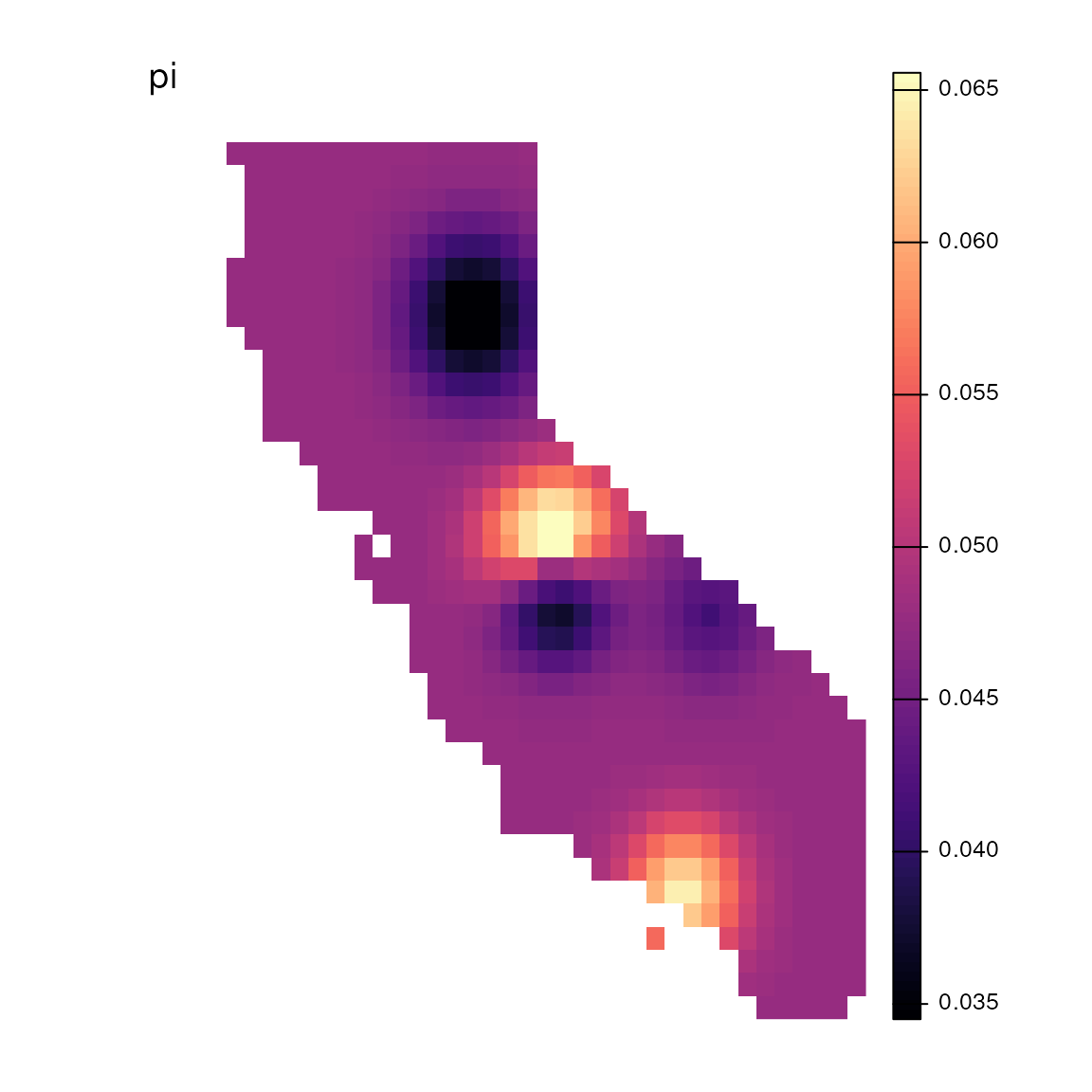
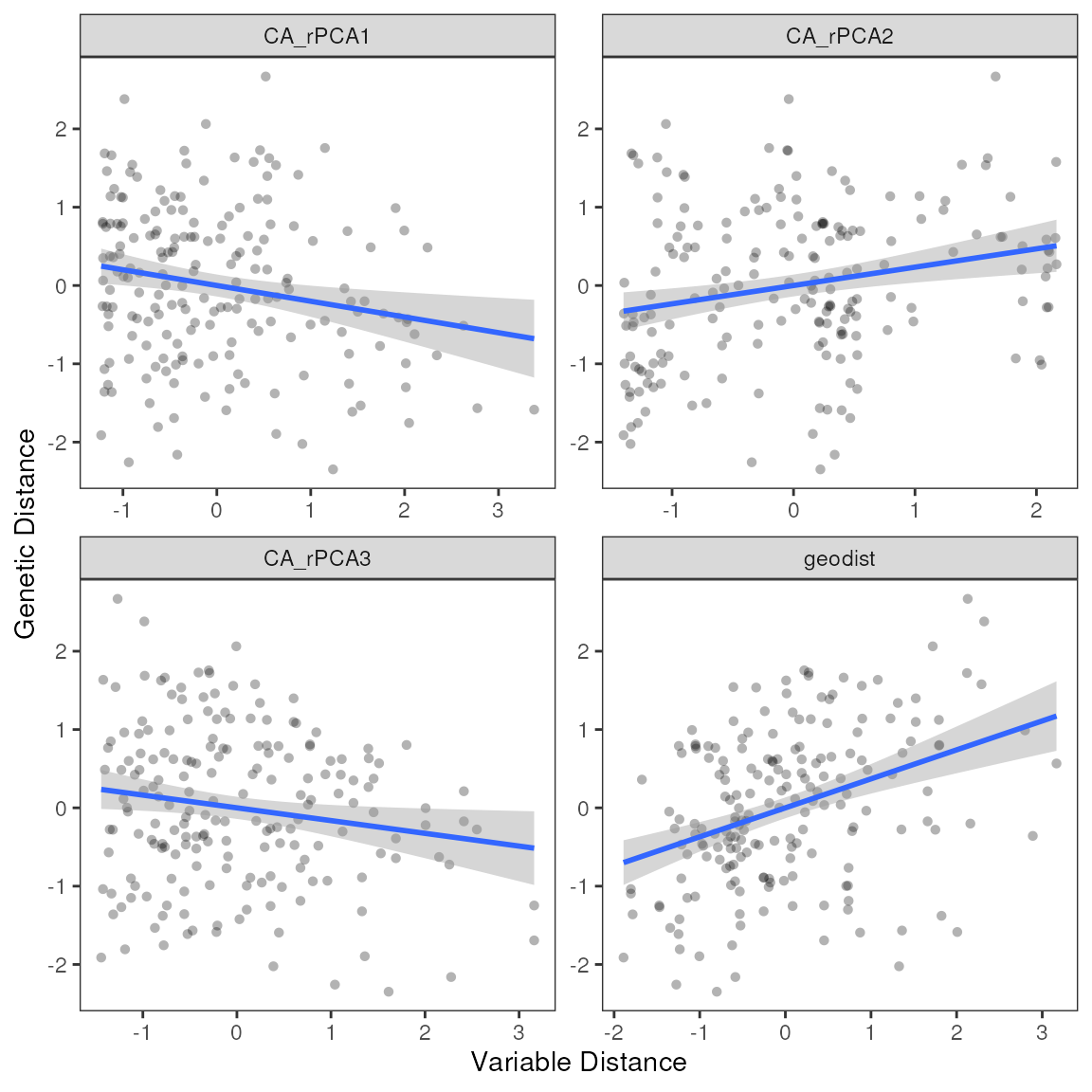
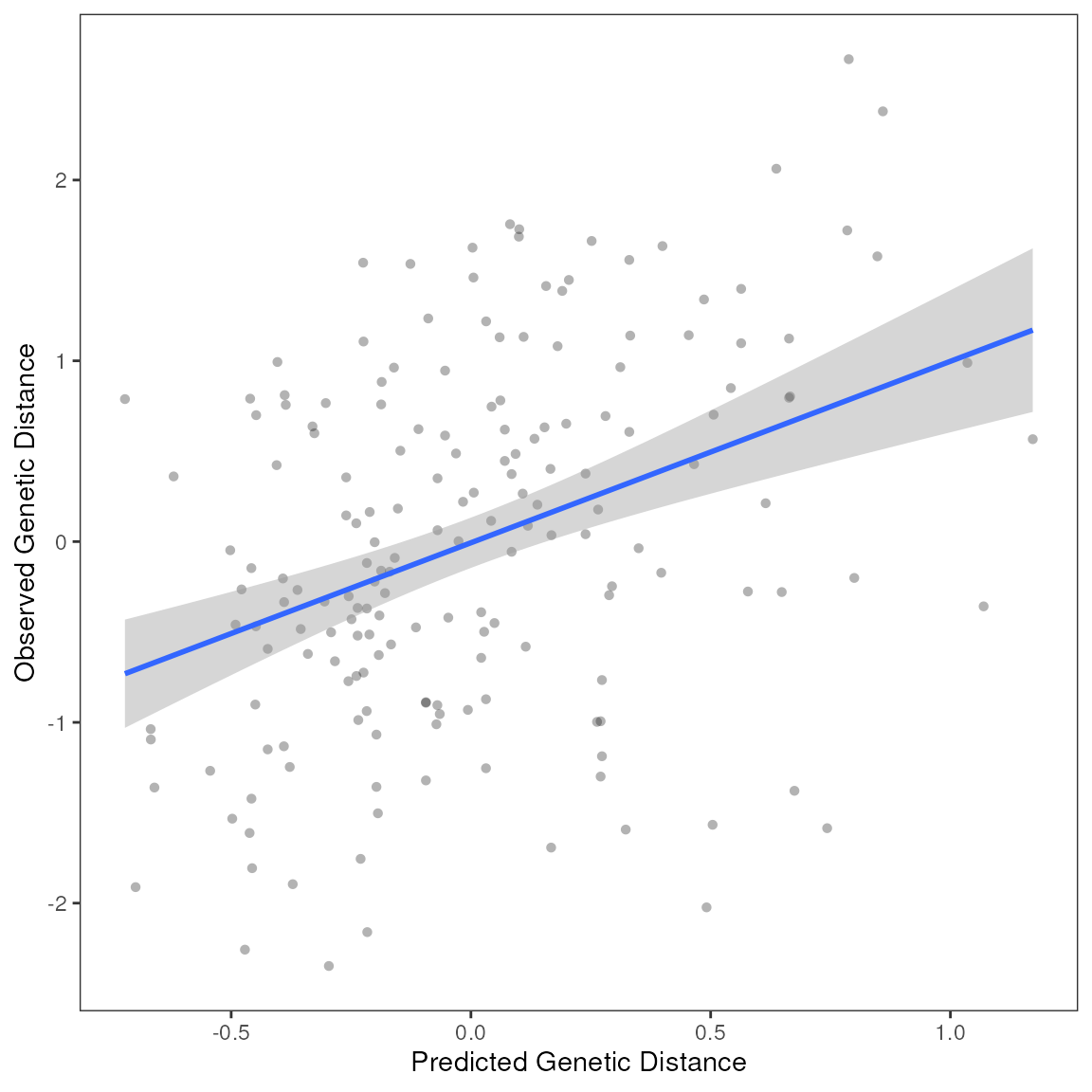
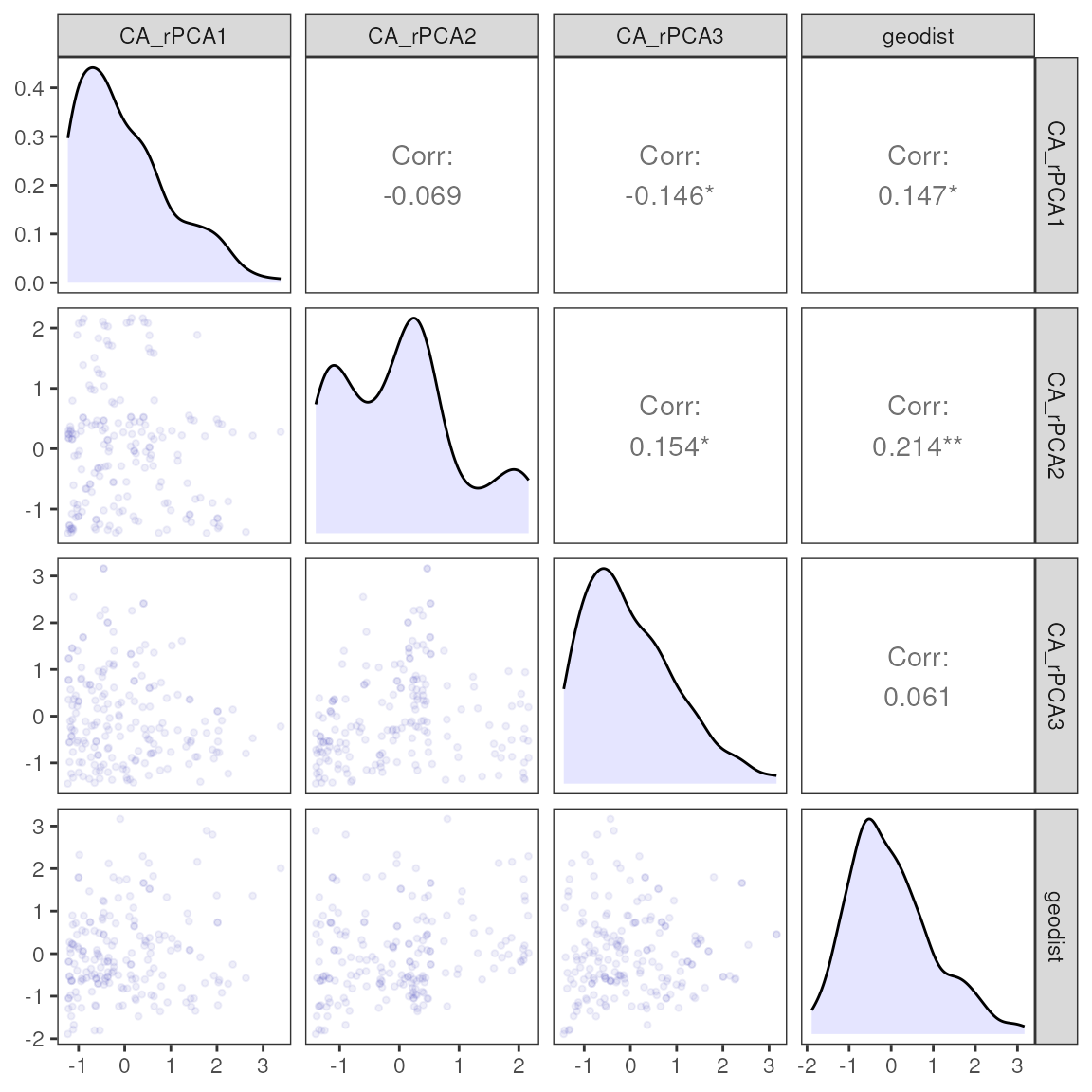
| var | estimate | p | 95% Lower | 95% Upper |
|---|---|---|---|---|
| geodist | 0.37 | 0.01 | 0.26 | 0.48 |
| Intercept | 0.00 | 0.53 | −0.11 | 0.11 |
| R-Squared: | 0.14 |
|
|
|
| F-Statistic: | 29.83 |
|
|
|
| F p-value: | 0.01 |
|
|
|
| predictor | coefficient |
|---|---|
| Geographic | 0.66 |
| CA_rPCA1 | 0.00 |
| CA_rPCA2 | 0.31 |
| CA_rPCA3 | 0.00 |
| % Explained: | 24.521 |
| 1 The percentage of null deviance explained by the fitted GDM model. | |
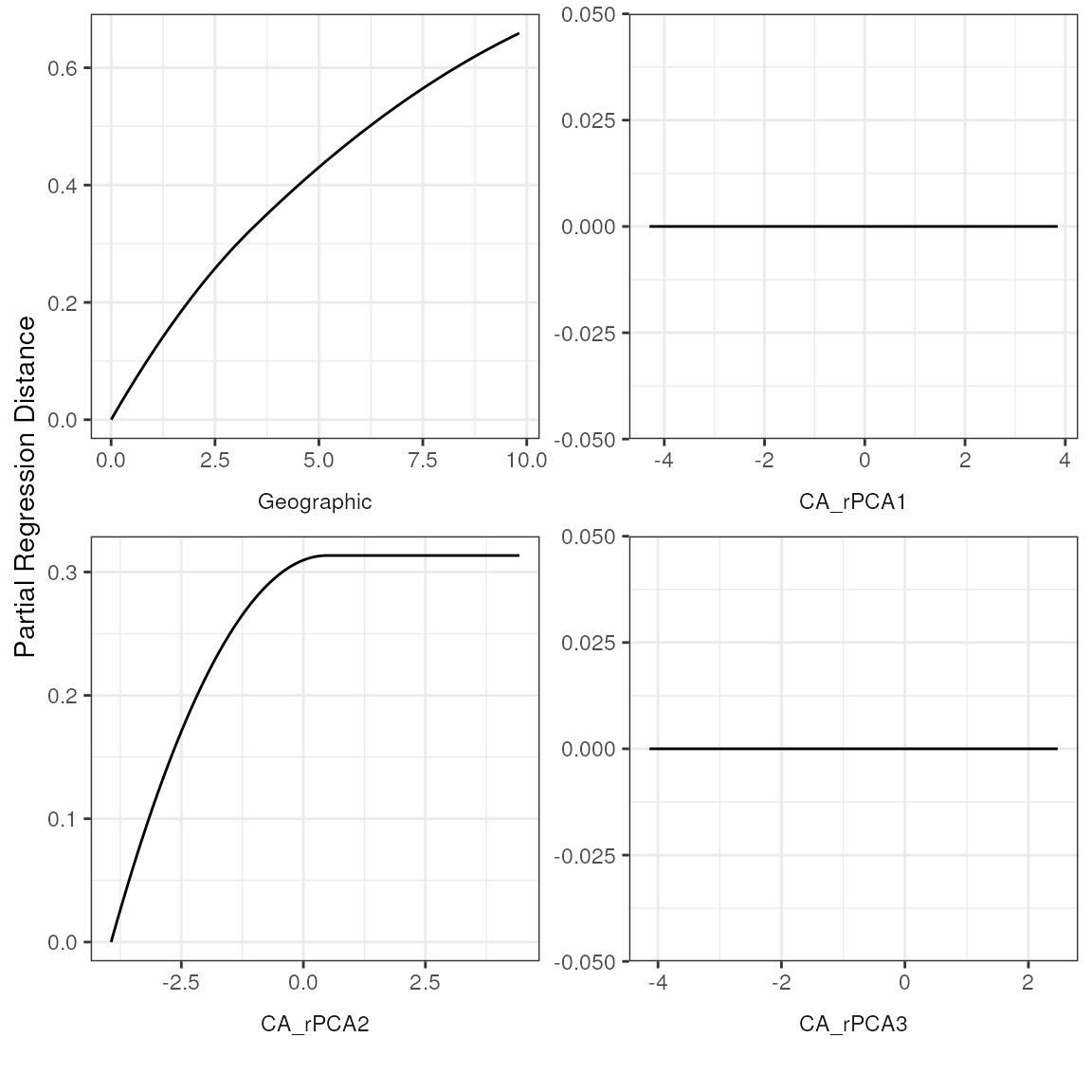
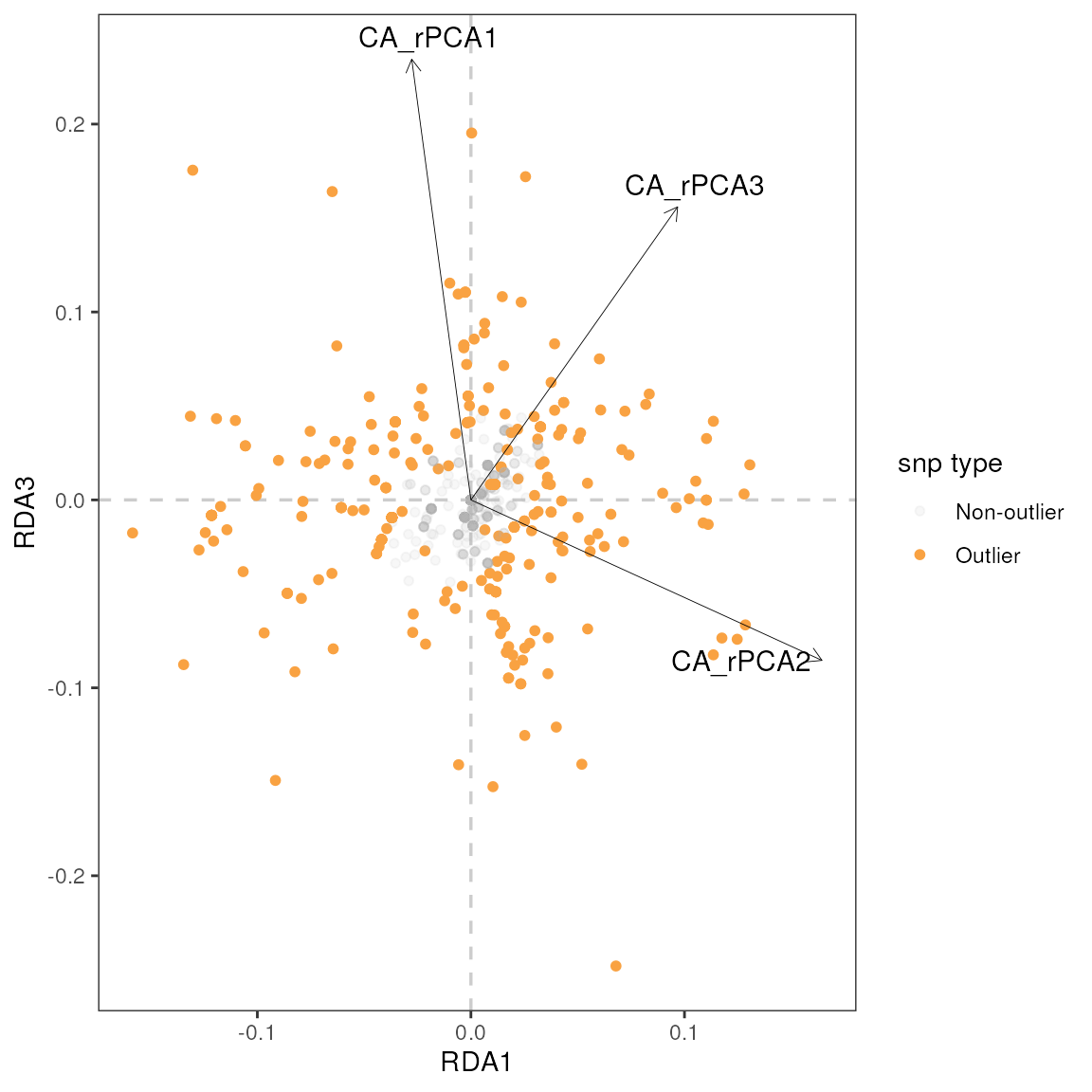
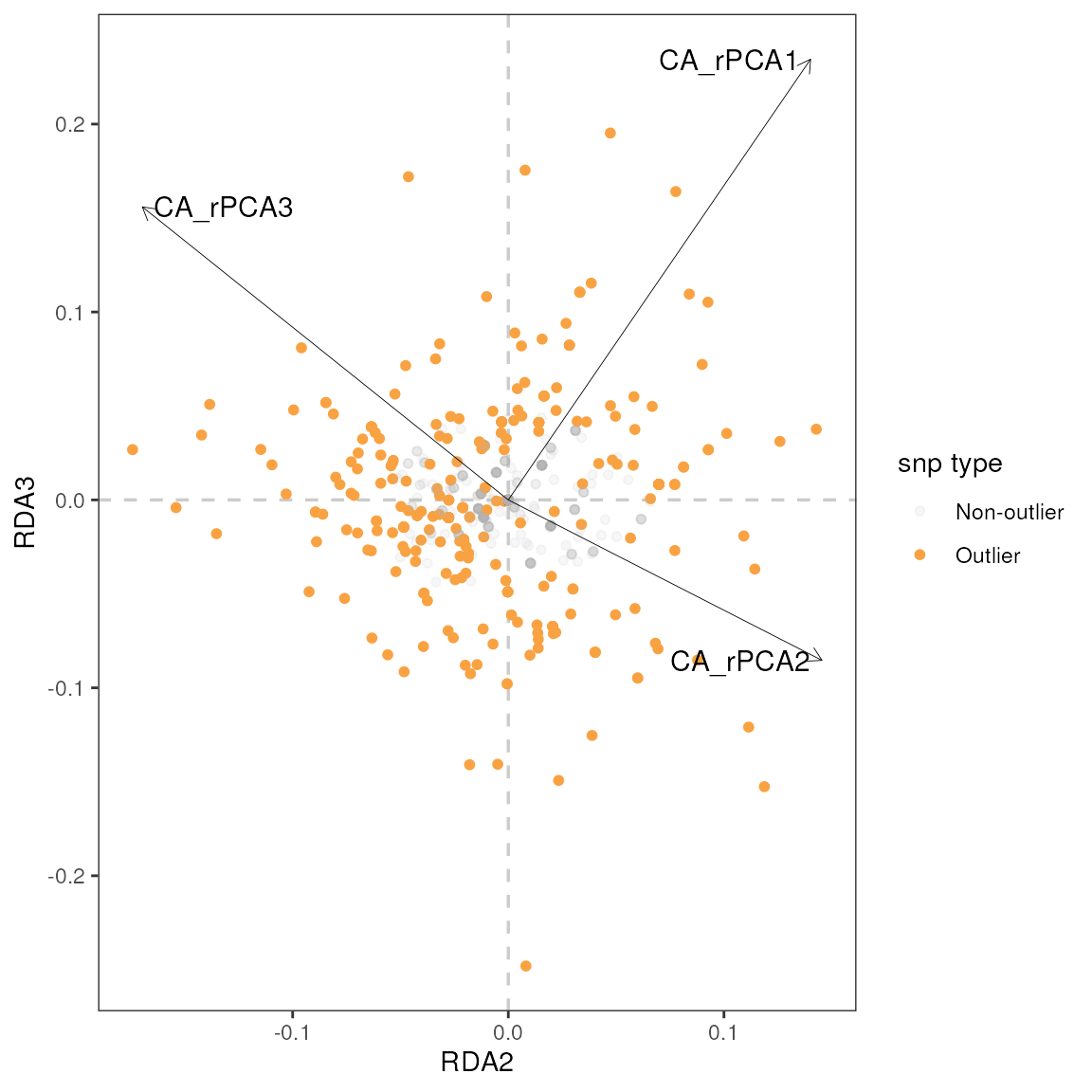
| r | p | snp | var |
|---|---|---|---|
| 0.61 | 0.00 | Locus_1841 | CA_rPCA3 |
| -0.58 | 0.00 | Locus_1319 | CA_rPCA2 |
| -0.58 | 0.00 | Locus_2394 | CA_rPCA2 |
| -0.53 | 0.00 | Locus_1247 | CA_rPCA2 |
| -0.52 | 0.01 | Locus_2733 | CA_rPCA2 |
| -0.51 | 0.01 | Locus_2602 | CA_rPCA2 |
| -0.49 | 0.01 | Locus_3143 | CA_rPCA2 |
| -0.48 | 0.01 | Locus_2665 | CA_rPCA3 |
| 0.47 | 0.01 | Locus_350 | CA_rPCA3 |
| 0.46 | 0.01 | Locus_2400 | CA_rPCA3 |
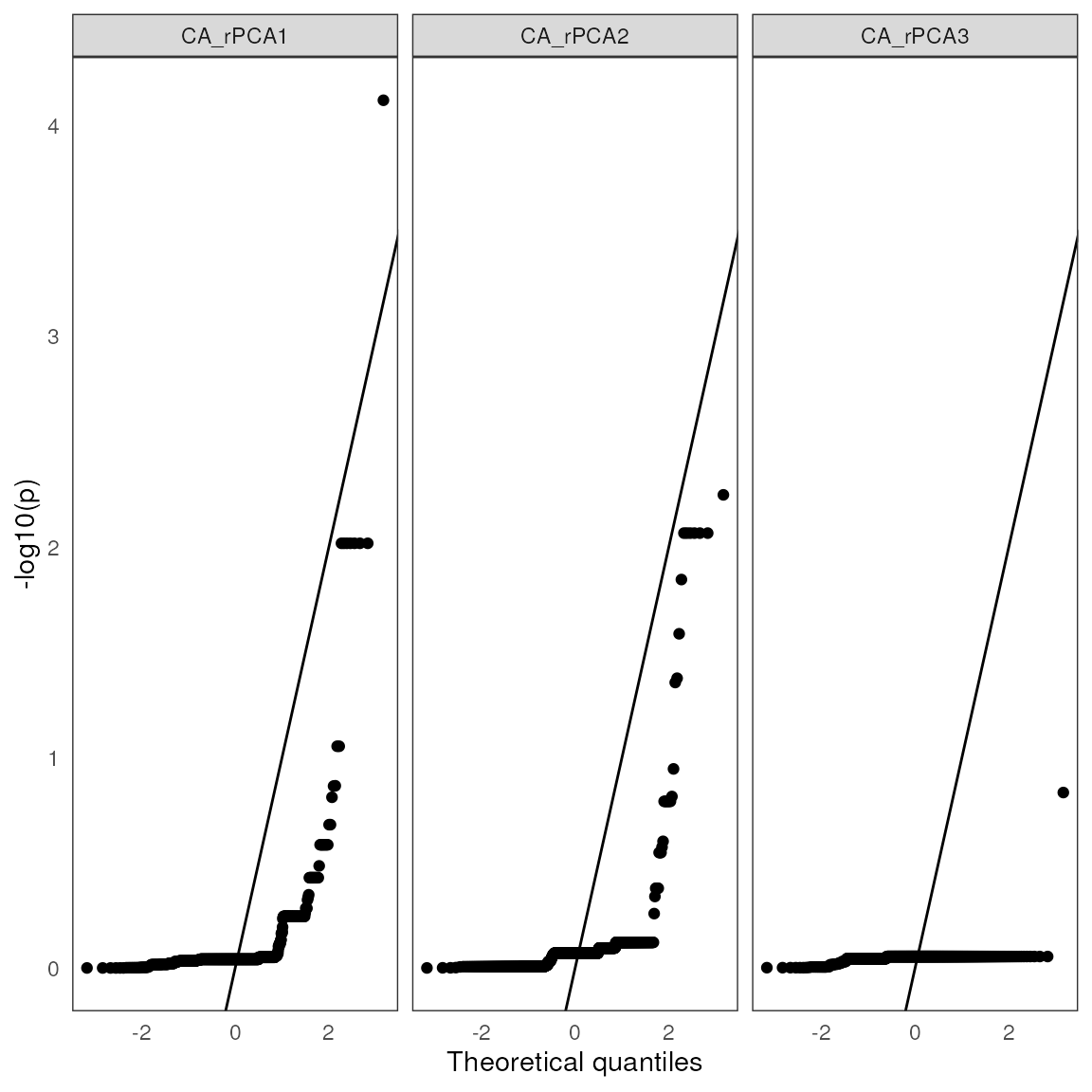
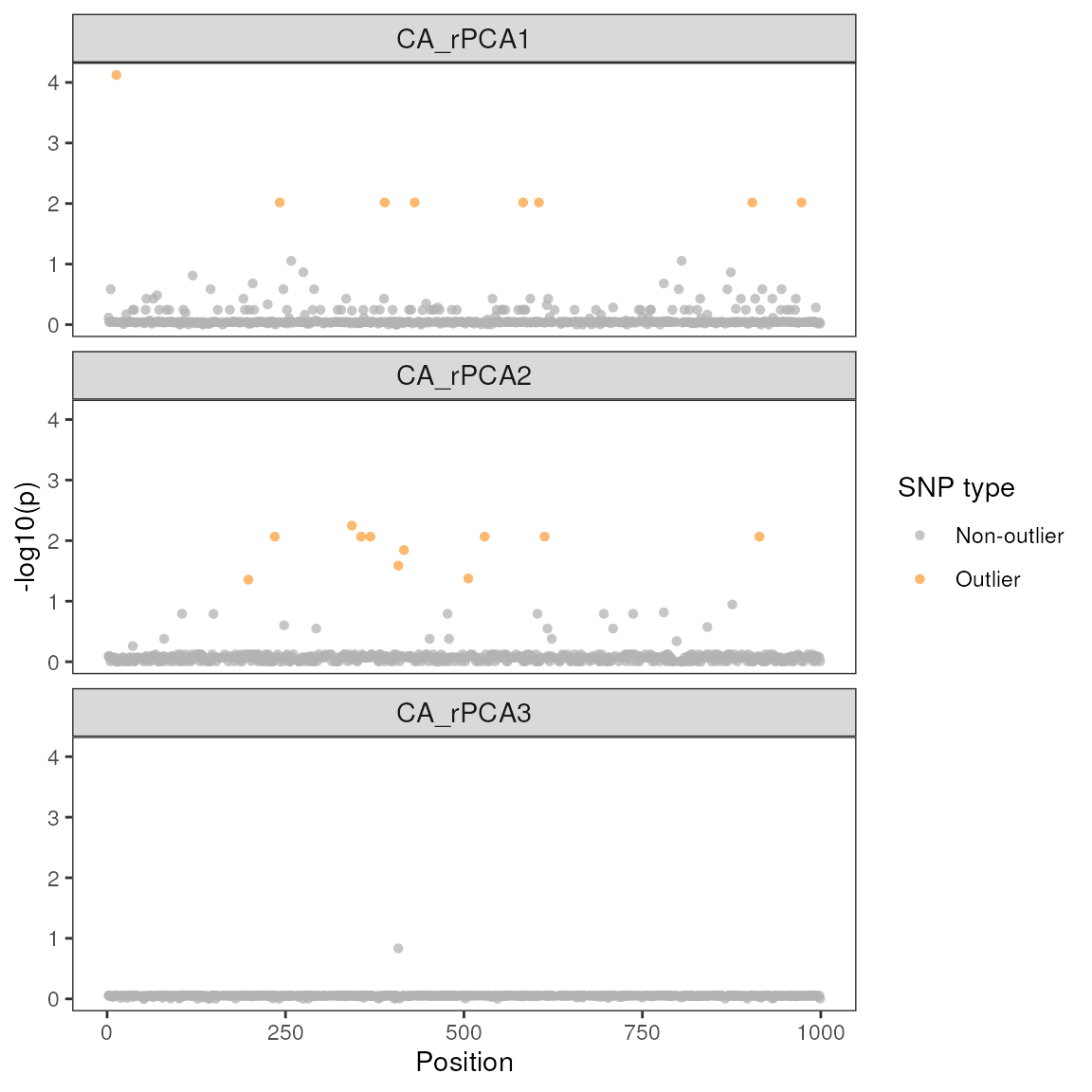
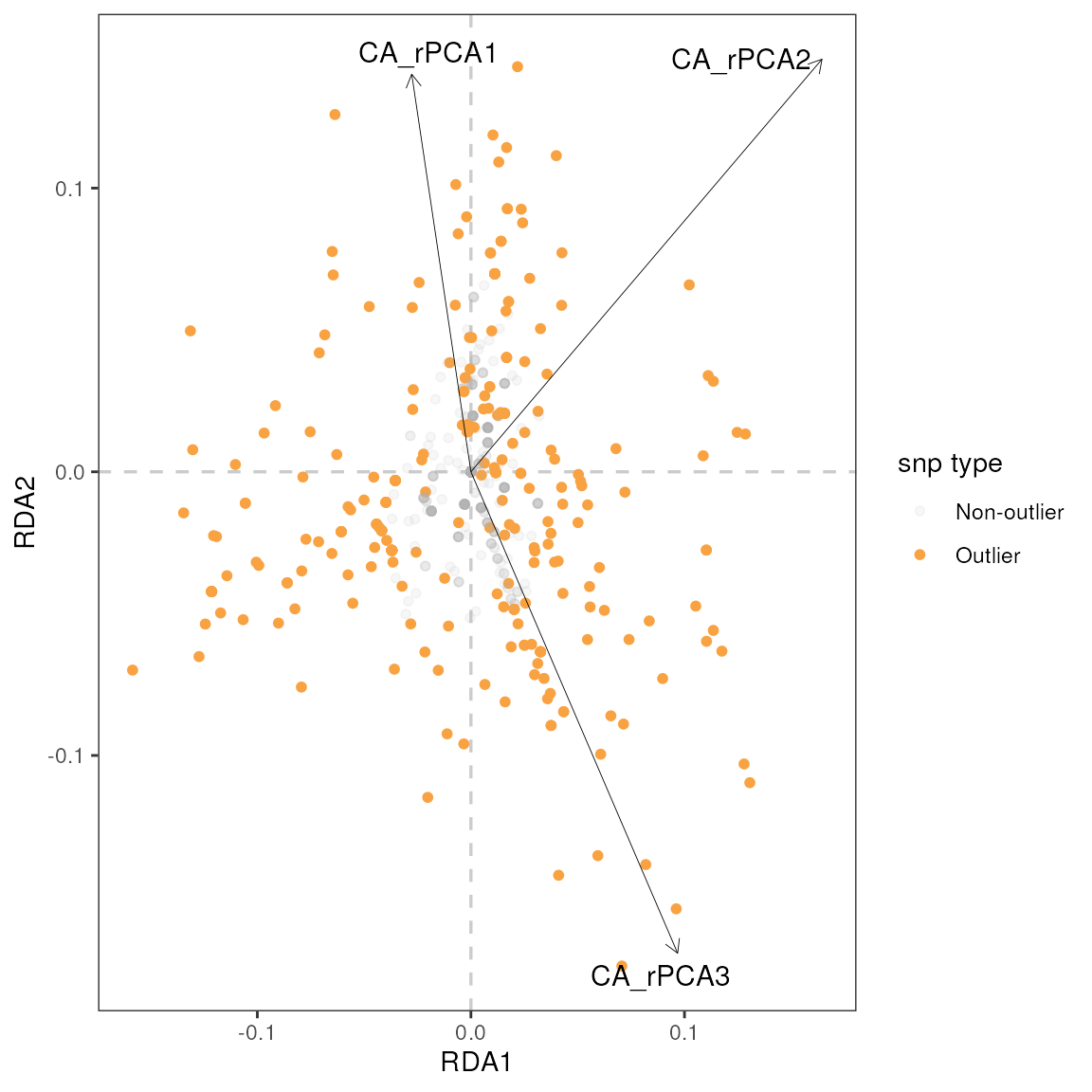
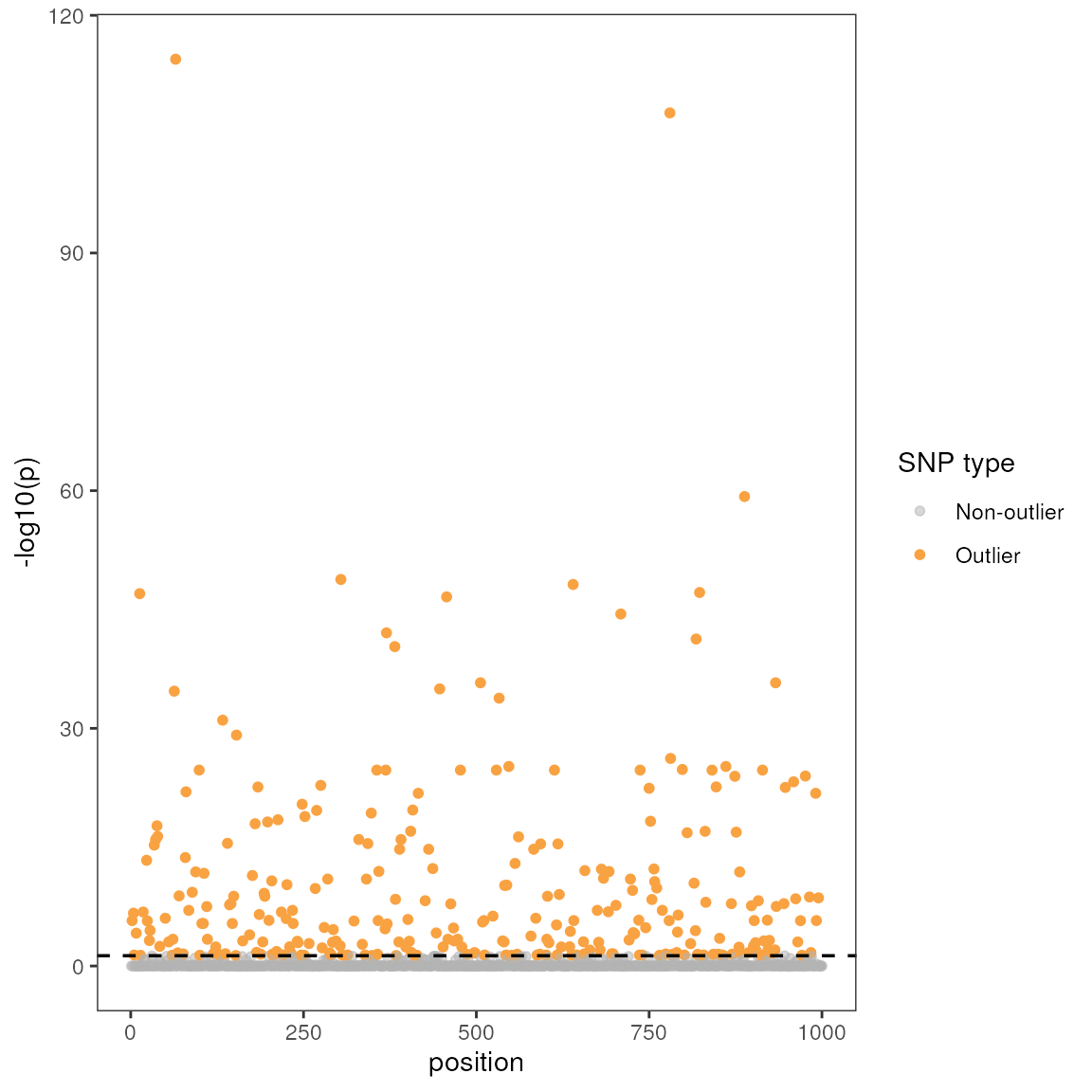
| snp | variable | B1 | z-score | p-value | calibrated z-score | calibrated p-value | adjusted p-value |
|---|---|---|---|---|---|---|---|
| Locus_90 | CA_rPCA1 | 0.24 | 4.36 | 0 | 28.07 | 0 | 0.00 |
| Locus_1278 | CA_rPCA2 | -0.21 | -4.89 | 0 | 15.27 | 0 | 0.01 |
| Locus_1318 | CA_rPCA2 | -0.21 | -4.89 | 0 | 15.27 | 0 | 0.01 |
| Locus_1801 | CA_rPCA2 | -0.21 | -4.89 | 0 | 15.27 | 0 | 0.01 |
| Locus_2045 | CA_rPCA2 | -0.21 | -4.89 | 0 | 15.27 | 0 | 0.01 |
| Locus_2947 | CA_rPCA2 | -0.21 | -4.89 | 0 | 15.27 | 0 | 0.01 |
| Locus_1724 | CA_rPCA2 | -0.20 | -4.27 | 0 | 11.62 | 0 | 0.04 |
| Locus_1247 | CA_rPCA2 | -0.19 | -5.57 | 0 | 19.76 | 0 | 0.01 |
| Locus_1477 | CA_rPCA2 | 0.17 | 4.70 | 0 | 14.06 | 0 | 0.01 |
| Locus_786 | CA_rPCA2 | -0.17 | -4.22 | 0 | 11.36 | 0 | 0.04 |
| 1 LFMM effect size | |||||||
The do_everything_for_me() function returns a list with
each analysis’s results as objects named according to the method. These
formats are identical to those obtained when running respective
“do_everything” functions (e.g., tess_do_everything()
function output is identical to that within the tess
object).
For methods with model selection (e.g., GDM and MMRR), the default of
do_everything_for_me() is to run with model selection
("best"), and if no significant variables are found, the
function will revert to running the "full" model for these
methods.